AI-aided Analysis of Retinal Fluorescent Biomarker
In collaboration with Novai, our engineers applied machine learning and image analysis techniques to clinical ophthalmology. We developed a modular system capable of detecting and classifying cellular glaucoma biomarkers in low-resolution retinal images. This approach reliably identifies early signs of the disease even with limited image quality and can be easily adapted to new datasets.

 Business Challenge
Business Challenge
Clinical ophthalmology is one of the many fields where image-related diagnostic techniques offer insights into eye diseases, using medical datasets. Machine Learning (ML) has met with great success in solving many visual and auditory recognition tasks.
ML allows for comprehensive, fast, and non-invasive analysis of digital data. The medical imaging field uses ever-increasing computing power and cloud storage, as well as novel algorithms and methods of data generation. The growing potential of ML to automatically detect, classify, identify and verify specific features in ocular diseases will empower practitioners with highly intelligent tools and high-quality diagnosis, and will further improve personalized healthcare services. Applying AI methods to medical tasks allows for ensuring medical data safety and efficient usage. Collaborations between medical imaging and AI disciplines have proven highly effective in many fields, where conventional diagnostic methods greatly depend on the experience and knowledge of physicians, which cannot be easily scaled and extended.
Our partner is Novai, a digital biomarker startup that has developed DARC (Detection of Apoptosing Retinal Cells) technology, which combines an innovative patented biologic with advanced artificial intelligence algorithms to determine disease activity in the retina at the cellular level. The main task was to leverage Novai’s high-resolution CNN-based algorithm for biomarker characterization to apply to low-resolution images.
 Solution Overview
Solution Overview
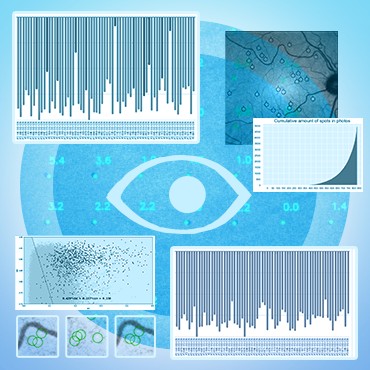
The main problem of diagnosing glaucoma at the different stages of the disease progression can be efficiently reduced by the detection of specific biomarkers on images of the patient’s retina. This approach also has the additional benefit of multiplying the data points for the training of ML classification methods, as the number of markers (spots) is much bigger than the number of eye images.
The high-level pipeline for the solution included several stages:
- dataset verification and clean-up
- image registration
- search for candidate spots
- filtering of candidates
- binary classification of the spots
At the final stage, the selected candidates are fed into a custom-trained convolutional neural network (an ensemble of MobileNet-architecture CNN's proved to be a sufficiently balanced choice, allowing for fast re-training on new data, efficient production execution, and provide good target accuracy). The solution is now being tested and implemented in health care institutions.
 Technical Details
Technical Details
The first step was to check the correctness of the data provided by the partners, eliminate duplicates, and determine the consistency of the data. The same images were marked by several experts, as a result of which, quite often, different eye defects (visible spots, which are markers of eye disease) were noted on various versions of the marked image. To do this, we used the SimpleITK library - “Insight Segmentation and Registration Toolkit” (which interface with most image processing filters allows you to perform image analysis workflows with a convenient syntax). This library is well-established for medical image analysis. Many medical research labs include SimpleITK as a key building block because it provides a wide range of image filtering and image input/output components with a user-friendly interface. In our pipeline, a python script is launched first, that forms an object with which the program then works, highlighting the necessary components. In the first stage, the first affine transformation is performed with the alignment of the geometric centers of the two images and the construction of the registration instance.
Then the metric type and mask are set. The optimizer uses the gradient descent method. After the optimizer's work is completed, the result of the work is generated and transferred to the second BSpline transformation, which is performed similarly to the affine one.
After that, the data is sent to the CNN implemented on the PyTorch framework to count the number of spots.
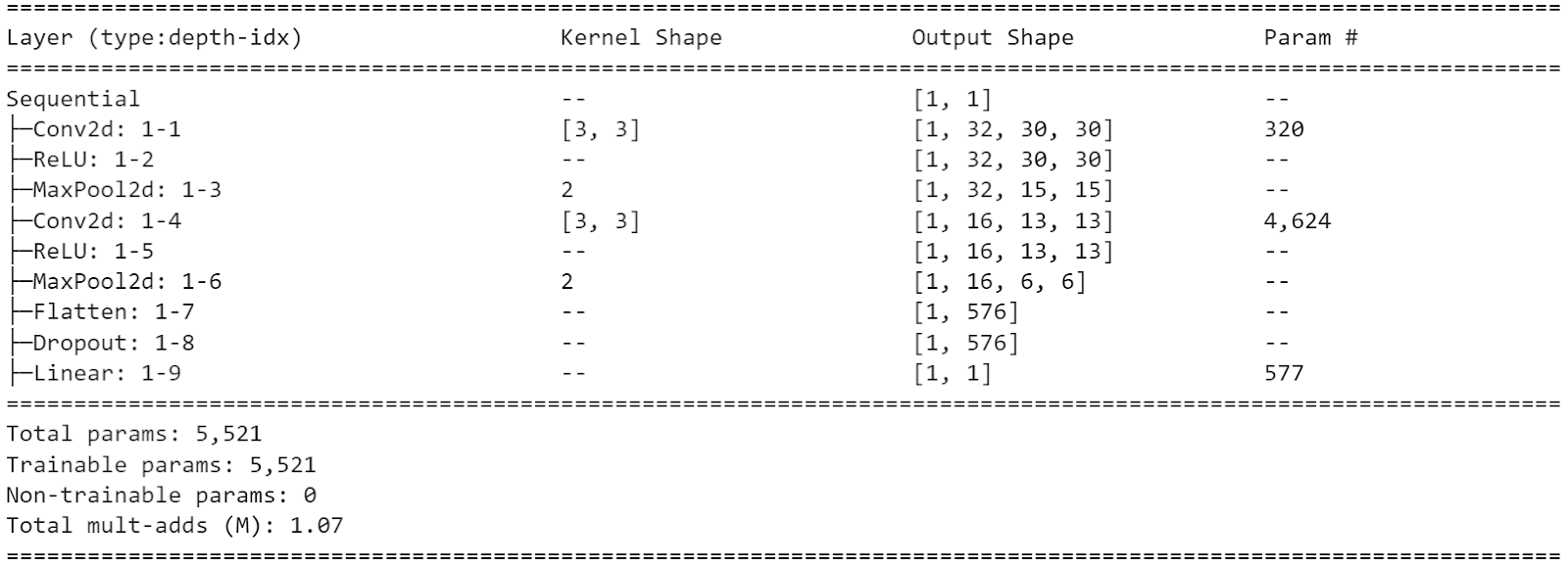
The CNN architecture with 5521 parameters is below:
Technology Stack

Python

SimpleITK

