Advantages of tree architectures over Convolutional Networks: A Performance Study
Traditionally, the methods of training solutions for deep learning (DL) have their roots in the principles of the human brain. In this context, neurons are represented as nodes connected to each other, and the strength of these connections changes as neurons interact. Deep neural networks consist of three or more layers of nodes, including input and output layers. However, these two learning scenarios are significantly different. Firstly, effective DL architectures require dozens of hidden feedforward layers, which are currently expanding to hundreds, while the brain's dynamics consist of just a few feedforward layers.
Secondly, deep learning architectures typically include many hidden layers, with the majority of them being convolutional layers. These convolutional layers search for specific patterns or symmetries in small sections of input data. Then, when these operations are repeated in subsequent hidden layers, they help identify larger features that define the class of input data. Similar processes have been observed in our visual cortex, but approximated convolutional connections have been mainly confirmed from the retinal input to the first hidden layer.
Another complex aspect of deep learning is that the backpropagation technique, which is crucial for the operation of neural networks, has no biological analogue. This method adjusts the weights of neurons so that they become more suitable for solving the task. During training, we provide the network with data input and compare how much it deviates from what we would expect. We use an error function to measure this difference.
Then we begin to update the weights of neurons to reduce this error. To do this, we consider each path between the input and output of the network and determine how each weight on this path contributes to the overall error. We use this information to correct the weights.
Convolutional and fully connected layers of the network play a crucial role in this process, and they are particularly efficient due to parallel computations on graphics processing units. However, it is worth noting that such a method has no analogs in biology and differs from how the human brain processes information.
So, while deep learning is powerful and effective, it is an algorithm developed exclusively for machine learning and does not mimic the biological learning process.
Researchers at Bar-Ilan University in Israel wondered if it's possible to develop a more efficient form of artificial intelligence by employing an architecture resembling an artificial tree. In this architecture, each weight has only one path to the output unit. Their hypothesis is that such an approach may lead to higher accuracy in classification than in more complex deep learning architectures that use more layers and filters. The study is published in the journal Scientific Reports.
The core of this study explores whether learning within a tree-like architecture, inspired by dendritic trees, can attain results as successful as those typically achieved using more structured architectures involving multiple fully connected and convolutional layers.
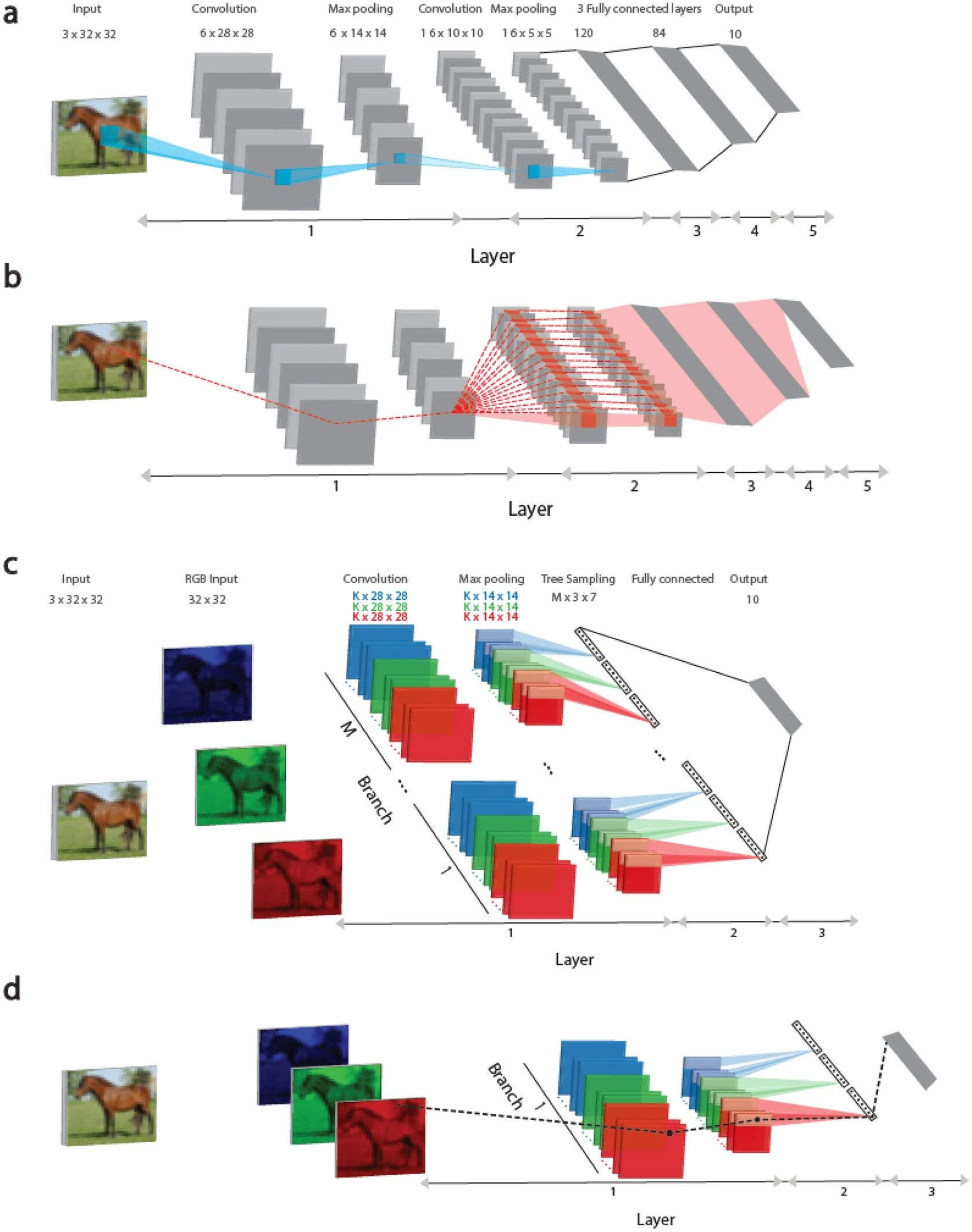
This study presents a learning approach based on tree-like architectures, where each weight is connected to the output unit by only one route, as shown in Figure 1 (c, d). This approach represents a step closer to implementing biological learning realistically, considering recent findings that dendrites (parts of neurons) and their immediate branches can change, enhancing the strength and expressiveness of signals passing through them.
Here, it is demonstrated that the performance metrics of the proposed Tree-3 architecture, which has only three hidden layers, outperform the achievable success rates of LeNet-5 on the CIFAR-10 database.
In Figure 1 (a), the convolutional architectures of LeNet-5 and Tree-3 are considered. The LeNet-5 convolutional network for the CIFAR-10 database consists of RGB input images sized 32 × 32, belonging to 10 output labels. The first layer consists of six (5 × 5) convolutional filters, followed by (2 × 2) max-pooling. The second layer consists of 16 (5 × 5) convolutional filters, and layers 3–5 have three fully connected hidden layers of sizes 400, 120, and 84, which are connected to 10 output units.
In Figure 1 (b), the dashed red line indicates the scheme of routes influencing the weight updates belonging to the first layer on panel (a) during the error backpropagation technique. A weight is connected to one of the output units by multiple routes (dashed red lines) and can exceed one million. It is important to note that all weights at the first layer are equated to the weights of 6 × (5 × 5), belonging to the six convolutional filters, as shown in Figure 1 (c).
The Tree-3 architecture consists of M = 16 branches. The first layer of each branch consists of K (6 or 15) (5 × 5) filters for each of the three RGB channels. Each channel is convolved with its own set of K filters, resulting in 3 × K different filters. The convolutional layer filters are the same for all M branches. The first layer concludes with max-pooling consisting of non-overlapping (2 × 2) squares. As a result, there are (14 × 14) output units for each filter. The second layer consists of a tree-like (non-overlapping) sampling (2 × 2 × 7 units) of K-filters for each RGB color in each branch, resulting in 21 output signals (7 × 3) for each branch. The third layer fully connects the outputs of 21 × M branches of layer 2 to 10 output modules. ReLU activation function is used for online learning, while Sigmoid is used for offline learning.
In Figure 1 (d), the dashed black line marks the scheme of one route connecting the updated weight at the first layer, as depicted in Figure 1 (c), during the error backpropagation technique to the output device.
To solve the classification task, researchers applied the cross-entropy cost function and utilized the stochastic gradient descent algorithm to minimize it. To fine-tune the model, optimal hyperparameters such as learning rate, momentum constant, and weight decay coefficient were found. To evaluate the model, several validation datasets consisting of 10 000 random examples, similar to the test dataset, were used. The average results were calculated, considering the standard deviation from the stated average success metrics. Nesterov's method and L2 regularization were applied in the study.
Hyperparameters for offline learning, including η (learning rate), μ (momentum constant), and α (L2 regularization), were optimized during offline learning, which involved 200 epochs. Hyperparameters for online learning were optimized using three different dataset example sizes.
As a result of the experiment, an effective approach to training a tree-like architecture, where each weight is connected to the output unit by only one route, was demonstrated. This approximation to biological learning and the ability to use deep learning with greatly simplified dendritic trees of one or several neurons. It is important to note that adding a convolutional layer to the input helps preserve the tree-like structure and improve success compared to architectures without convolution.
While the computational complexity of LeNet-5 was notably higher than that of the Tree-3 architecture with comparable success rates, its efficient implementation necessitates new hardware. It is also expected that training the tree-like architecture will minimize the likelihood of gradient explosions, which is one of the main challenges for deep learning. Introducing parallel branches instead of the second convolutional layer in LeNet-5 improved success metrics while maintaining the tree-like structure. Further investigation is warranted to explore the potential of large-scale and deeper tree-like architectures, featuring an increased number of branches and filters, to compete with contemporary CIFAR-10 success rates. This experiment, using LeNet-5 as a starting point, underscores the potential benefits of dendritic learning and its computational capabilities.
