Stochastic cluster embedding – a new method for visualizing large datasets
An amazing feature of the human brain is the ability to find differences even in a huge amount of visual information. When studying large amounts of data, this ability turns out to be very useful, because the content of the data must be compressed into a form understandable to human intelligence. For visual analytics, the problem of dimensionality reduction remains the main one.
Scientists from Aalto University and the University of Helsinki at the Finnish Center for Artificial Intelligence (FCAI) conducted a study where they tested the functionality of the most well-known visual analytics methods and found that none of them work when the volume of data increases significantly. For example, the t-SNE, LargeViz, and UMAP methods could no longer distinguish extremely strong signal groupings of observations in the data, when the number of observations runs into the hundreds of thousands. The t-SNE, LargeViz, and UMAP methods no longer work properly.
The researchers have developed a new non-linear dimensionality reduction method called Stochastic Cluster Embedding (SCE) for better cluster visualization. It aims to visualize data sets as clearly as possible and is designed to visualize data clusters and other macroscopic features in such a way that they are as distinct, easy to observe and human-understandable as possible. SCE uses graphics acceleration similar to modern artificial intelligence methods for computing in neural networks.
The discovery of the Higgs boson was the basis for the invention of this algorithm. The data set for the experiments associated with it contains over 11 million feature vectors. And these data required convenient, clear visualization. This inspired the scientists to develop a new method.
The researchers generalized the SNE using a family of I-divergences, parameterized by a scale factor s, between non-normalized similarities in input and output space. SNE is a special case in the family where s is chosen as the normalizing factor for similarity of outputs. However, during testing, it was found that the best value of s for cluster visualization often differs from the value chosen by the SNE. Therefore, to overcome the shortcoming of t-SNE, the new SCE method uses a different approach that mixes input similarities when calculating s. The coefficient is adaptively adjusted when optimizing the new learning objective and thus the data points are better clustered. The researchers also developed an efficient optimization algorithm using asynchronous stochastic descent over block coordinates. The new algorithm can use parallel computing devices and is suitable for mega-scale tasks with large amounts of data.
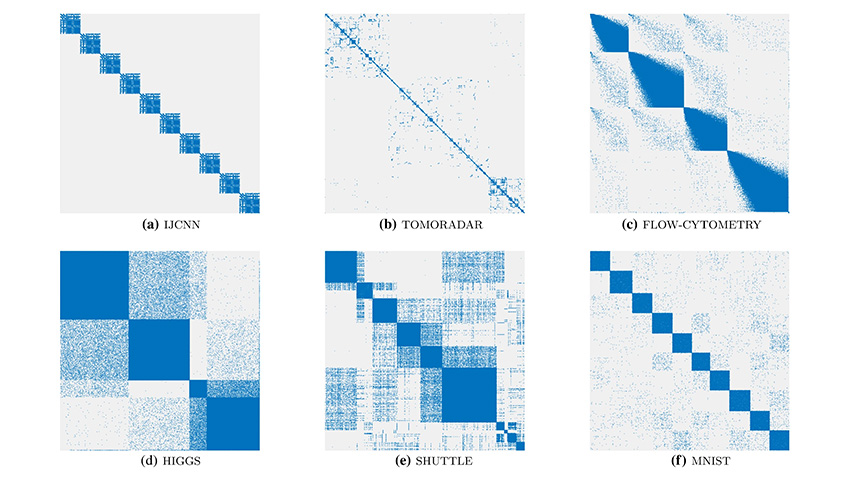
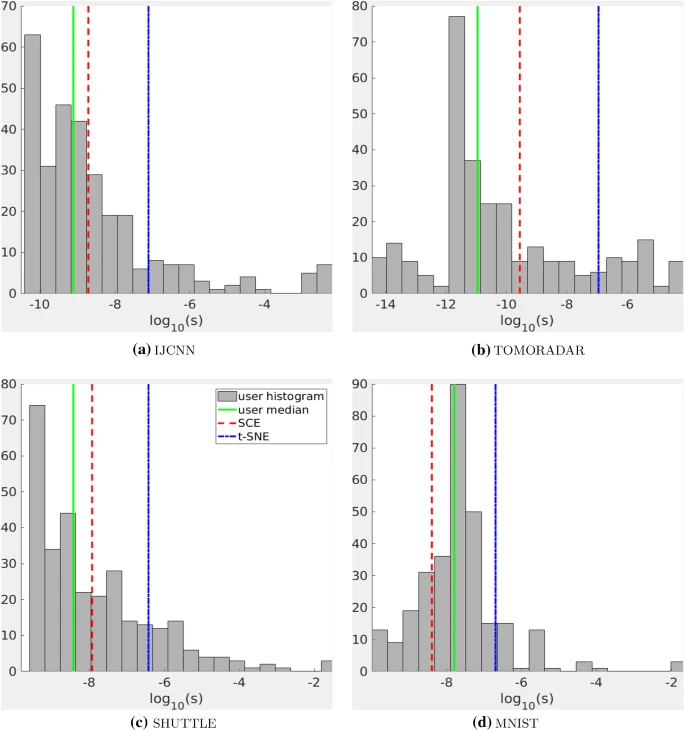
During the development of the project, the scientists tested the method on various sets of real data and compared it with other modern NLDR methods. Users participating in the testing selected the most appropriate visualizations that matched the range of s values for viewing clusters. The researchers then compared the resulting s values in SCE and t-SNE to see which was closer to human choice. The four smallest datasets IJCNN, TOMORADAR, SHUTTLE and MNIST were used for testing. For each dataset, test participants were presented with a series of visualizations where they used a slider to indicate an s value and tested the corresponding precomputed visualization. The user chose the preferred value of s for cluster visualization.
The test results clearly demonstrate that the s chosen by SNE is to the right of the human median (solid green line) for all datasets. This suggests that for humans, GSNE with smaller s is often better than t-SNE for cluster visualization. In contrast, the SCE selection (red dashed lines) is closer to the human median for all four datasets.
By applying the Stochastic Cluster Embedding method to data on the Higgs boson, their most important physical characteristics were clearly identified. The new non-linear dimensionality reduction method Stochastic Clustering Embedding for better cluster visualization works several orders of magnitude faster than previous methods, and is also much more reliable in complex applications. It modifies t-SNE using an adaptive and efficient compromise between attraction and repulsion. Experimental results demonstrated that the method can consistently identify internal clusters. In addition, scientists have provided a simple and fast optimization algorithm that can be easily implemented on modern parallel computing platforms. Efficient software has been developed that uses asynchronous stochastic block gradient descent to optimize a new family of objective functions. Experimental results have shown that the method consistently and significantly improves the visualization of data clusters compared to modern stochastic Neighbor Embedding approaches.
The code of the method is publicly available at github.
